Professor Andrew Baldwin

Andrew Baldwin
Professor of Physical and Theoretical Chemistry
Contact
andrew.baldwin@chem.ox.ac.uk
01865 613200
Research group
Our research focuses on using biomolecular NMR techniques to study the biophysical basis of function, and malfunction, of proteins in health and disease. We combine advanced solution and solids NMR spectroscopy techniques with complementary methods such as ion-mobility mass spectrometry and electron microscopy in order to study the structure and dynamics of proteins, and relate this to their behaviour in the cell.
We have three major inter-twined research areas.
The biophysical basis of protein mis-folding diseases
A growing number of human disorders including Alzheimer’s disease (AD) and Parkinson’s disease (PD), type II diabetes and the prion diseases are associated with the deposition of proteinaceous ‘amyloid’ aggregates in human tissues. The incidence of these conditions is increasing as populations age, with an associated national cost to the UK alone larger than both cancer and heart disease combined.
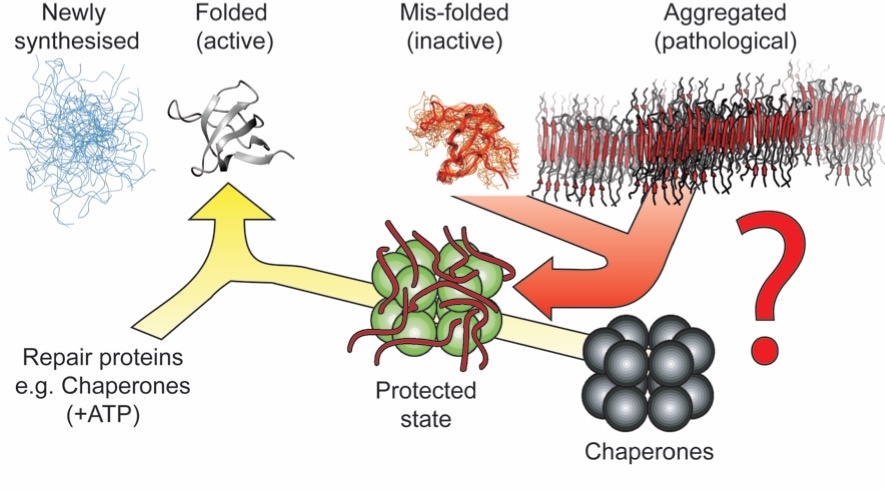
All is not lost however! A set of human 'molecular chaperone' proteins have the ability to prevent aberrant aggregation in vitro and are up-regulated and tightly associated with the tissues affected by these diseases in vivo. Our focus is to better understand the mechanism by which these proteins are able to inihibit, and in some cases prevent, protein aggregation and amyloid formation.
Experimentally, this is tremendously challenging. The systems that we need to study are incredibly heterogeneous, a property that confounds almost all high resolution techniques used by modern structural biology. Our approach is to combine cutting edge NMR spectroscopy, mass spectrometry and electron microscopy measurements. By doing so, we can characterize the structures and dynamics of these heterogeneous assemblies with very high precision.
The work of the group is therefore inherently inter-disciplinary. Novel experimental methodology and the computational techniques necessary to synthesize the information are developed in the group together with the molecular biology techniques required to make relevant high quality samples for detailed scrutiny.
Biomolecular NMR Spectroscopy: structure and dynamics at atomic resolution
Biological organisms rely on proteins to perform the majority of the chemical tasks needed to sustain life. All functions that proteins perform stem from the three-dimensional structures that they adopt. Because these structures are stabilized only by non-covalent interactions, at temperatures relevant to biology they are easily rearranged by thermal motion. Proteins are consequently very dynamic molecules that are best understood as an ensemble of inter-converting conformers, rather than single static structures.
The majority of experimental techniques available for structural biology however, report on the ‘average’ positions of the constituent atoms. In order to fully understand how biological molecules function, it is necessary to move beyond static structures. NMR spectroscopy is unique in that it can simultaneously describe the motions that proteins undergo as well as their structure. It can therefore yield great insight into how proteins ‘work’.
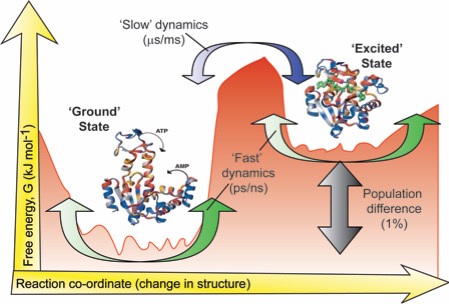
We develop and employ experiments to measure the movements a protein makes with unprecedented detail. Projects are available in both methods development, and in applying these novel techniques to a range of systems that are biochemically fascinating.
Translating atomic resolution information onto the oligomer level
The NMR experiments described above provide ‘local’ information about atomic structure and dynamics. We combine these data with measurements from ion-mobility mass spectrometry, which is able to give us three important bits of additional information: which oligomeric states are populated in solution, the timescale on which any quaternary structure rearranges and even the dimensions of individual oligomers, without the need for prior separation. By combining the local structural restraints derived from NMR, with the lower resolution but oligomer specific ‘global’ measurements, we are able to obtain a very rich picture of macromolecular behaviour.
In addition to pairing our NMR measurements with ion-mobility mass spectrometry, we also employ electron microscopy. The experimental triumphate of NMR, ion-mobility mass spectrometry and electron microscopy is proving itself to be incredibly powerful in studies of heterogeneous mixtures, such as those that spontaneously arise in problems associated with protein mis-folding disorders.
Andrew studied Natural Sciences at the University of Cambridge, as a member of Trinity College. He completed his PhD in the laboratory of Chris Dobson in the department of Chemistry, studying the biophysical properties of amyloid fibrils. He then moved to Toronto to develop novel NMR techniques for studying protein dynamics under Prof Lewis Kay, with EMBO and CIHR postdoctoral fellowships. This work was subsequently recognised by a presentation of a Harrison-Meldola medal in 2013. Since September 2012, Andrew has been part of Oxford Chemistry as both a BBSRC David Phillip’s fellow and a Fitzjames fellow of Merton College. He is currently Professor in Physical and Theoretical Chemistry, and a fellow of Pembroke College. He was awarded the 2016 BRSG-NMRDG NMR prize for ‘excellence in magnetic resonance’.


